I must begin by acknowledging my debt, both personal and intellectual, to Francisco Varela.
I met Francisco for the first time in 1991--at the very first European Conference on Artificial Life (ECAL). As co-organiser of the conference he was, of course, rather busy, so I was quite diffident in approaching him. Yet he was immediately welcoming, and happy to take the time to sit and discuss with me my initial tentative exploration of the concept of autopoiesis.
Subsequently we corresponded at some length, and in 1992 he accepted an invitation to Dublin to participate in the workshop Autopoiesis and Perception, which I co-organised with my colleague Noel Murphy [32]. We continued to correspond and meet at intervals over the following years. I particularly remember a discussion over dinner one evening during the third ECAL (Grenada, Spain, in 1995), where he dazzled me not only with his ability to maintain three simultaneous conversations with different people, but to do so in three (or more?) different languages, switching continuously between them!
In any case, that particular conversation laid the basis for a more intense correspondence over the following two years that culminated in an exhaustive (and exhausting?) re-examination of the original, computational model of autopoiesis [46]. This will be discussed in more detail below; for now it is enough to note that it ultimately resolved a number of difficult questions regarding the earlier work--but that this resolution was possible only because of Francisco's unfailing patience with my questions, and cheerful willingness to dredge both his memory and his files to locate critical contemporary records of the work.
Francisco Varela was a brilliant and original scientist. But my enduring memory is of Francisco the man: his enthusiasm, his infectious good humour, his idealism, and his sheer appetite for life. He is sorely missed.
What does it mean for something to be alive? Why are some things alive, and some not? Is there only one way of being alive, or many different and distinct ways?
These are the most fundamental questions which should define biology--and thus, in turn, Artificial Life--as a distinct field of enquiry. Yet the answers are not straightforward, and certainly not the subject of clear consensus. With the demise of vitalism, it was recognised that living systems are made of the same kinds of things as the non-living. It follows that the distinctiveness of living systems must arise from the particular manner or modes of organisation which they embody. But it has proved surprisingly difficult to articulate what this mode (or these modes) of living organisation consist in.
The project of autopoiesis was motivated inter alia by the attempt to answer just such questions. It proposes that, at some level, and in some way, all living things share a common organisation; and that this is characterised by two intimately entwined closure properties:
The central claim of autopoietic theory is that autopoietic organisation is necessary (though hardly sufficient) for the emergence of living phenomenology. If that claim is taken seriously, it naturally defines a particular programme within Artificial Life: namely the attempt to realise autopoietic organisation in artificial, especially computational, media. Indeed, the attempt to do just this has been an integral part of work on autopoiesis from its very inception. Accordingly, we will turn now to the historical setting and context for the formulation of autopoietic theory, and its seminal casting in a specifically computational context.
... the simulation rapidly provided the results our intuition had led us to expect: the spontaneous emergence in this artificial bi-dimensional world of units which self-distinguished by means of the formation of a `membrane', and which showed a capacity of self-repair. ... It is important to mention this article [46] here because it was the first publication on the idea of autopoiesis in English for an international public, which led the international community to take charge of the idea. In addition it anticipated what twenty years later would become the explosive field now called artificial life and cellular automata.
--Varela [45, p. 414]
As recounted by Varela, the concrete idea of autopoiesis arose only slowly, and in particular circumstances of time and place. The development seems to have originated around 1967 [45]. Varela was then a student (or ``apprentice'') under Humberto Maturana in the Department of Sciences at the University of Chile. Maturana was already well known, with an international reputation for his work on the neurophysiology of vision [see e.g., 13]; but he had started to develop a basic dissatisfaction with the idea of ``information'' as being key to understanding brain and cognition.
The problems here are profound, and are still by no means resolved. Nervous systems somehow contrive to imbue mere ``information'' with meaning and significance. Despite the continuing dominance of the ``information processing'' paradigm of cognition, such signification cannot be intrinsic to the raw information. Indeed, the very basis of the Shannon ``information theory'' is, precisely, to separate information from meaning. So signification is something that is somehow generated through the autonomous, self-referring, dynamics of a nervous system, constrained by its embedding in, and coupling with, the world it comes to know.
These ideas of autonomous circular organisation in nervous systems were developed by Maturana over the following several years. While Varela's personal trajectory took him to the USA at this time, the two continued to meet and collaborate together, and with Heinz von Foerster, in discussing these ideas. This culminated in the publication of Maturana's paper Neurophysiology of Cognition [22]. Varela retrospectively identified this as a critical step (to which, as Maturana acknowledged, both Varela and von Foerster had contributed)--``... the indisputable beginning of a turn in a new direction'' [45, p. 412]. In particular, it made the first connection between these issues of self-reference in nervous systems and corresponding, but even more basic, problems regarding the molecular constitution of the biological cell--which is to say, of life itself.
Varela returned to Chile in 1970, now as a colleague of Maturana in the Biology Department at the University of Chile. They deliberately focussed on exploring the nature of the organization of the living organism. At this point the problem was becoming clear as that of understanding or characterising the demarcation between living and non-living systems--but to do so in a way which was abstracted away from the specific biochemical contingencies of any particular form of life. So we see already here exactly the distinction between ``life as it is'' and ``life as it could be'' that eventually provided the intellectual foundation for the field of Artificial Life [17].
By mid-1971, the word itself, autopoiesis, had been coined, and Varela and Maturana had outlined a minimal model or exemplar. By this was meant an imaginary, abstract, chemical world, incorporating just those species and reactions that would suffice to allow the constitution of a minimal autopoietic system. This would therefore serve to isolate, in a very precise and concrete way, just what was, and was not, being pointed at by the new concept.
Thus, the minimal model served an important expository purpose--to give concrete form to what is otherwise a rather difficult and unfamiliar idea; but to do so in a way that strips away the many complex distractions that any real bio-chemical system would additionally present. Moreover, then, as now, no real living cell is yet described in sufficient detail to be able to definitively identify its constitution as autopoietic in any case.
Yet, the minimal model also served another, potentially much more important purpose: as a test of what had now become, not just a concept, but a theory of autopoiesis. Maturana and Varela conjectured that autopoietic organisation was capable of giving rise to characteristically biological phenomenology--in particular, to that most primitive cellular phenomenon of ``self-repair'', of the conservation of composite, macroscopic, cellular organisation even as the microscopic constituents of the cell are continuously turned over.
Ideally, of course, such a test case might have been implemented in ``real'', ``wet'', chemistry. But this is both technically challenging and, in any case, runs the risk of re-introducing many distracting complications that are not relevant to the restricted question being asked. So, instead, they hit on the idea of using a computer simulation; and, in particular, the idea of a low-level, fine-grained (``individual-'' or ``agent-based'') simulation.
This methodological idea was undoubtedly already ``in the air''. Varela himself described it as an ``obvious step'' [45, p. 414]. It drew explicitly on the pioneering work of John von Neumann--work carried out in the early 1950's, but which had been properly published only a few years prior to Varela and Maturana's work [47,29]; and on the seminal development of the Game of Life by Conway et. al., reported just as Varela and Maturana were already working along these lines [12].
On the other hand, some distinctive and highly original innovations were also required. The basic idea of a discrete, two-dimensional, space, with local interactions, clearly derives from von Neumann. However, Varela and Maturana introduced the concept of directly implementing (quasi-)stable ``particles'' which can move across the lattice, rather than relying on the relatively complex and fragile emergence of patterns of underlying node-states with this property (e.g., in the manner of gliders in the Game of Life). They also incorporated a deliberately stochastic dynamics, and inter-particle transformations or reactions--essential to anything motivated by the idea of realising an abstract ``chemistry'' (as opposed to an abstract ``physics''). From a contemporary perspective, writing in the pages of the Artificial Life Journal, these are now essentially standard techniques; but it is important to recognise the originality and insight that they represented in that much earlier era.
In any case, the assistance of Ricardo Uribe of the School of Engineering at the University of Chile was enlisted, and, as reported in the opening quote of this section, a computer simulation of the minimal model was created which ``rapidly'' confirmed the qualitative expectations of Varela and Maturana.
By the end of 1971 then, the theory of autopoiesis in its most basic form--as a theory of the molecular organisation of living cells--had been quite fully articulated, and tested in the form of a highly simplified (and therefore highly demanding) computer simulation. Two texts had been written: a comprehensive technical account, and a draft of a shorter summary article (including the simulation results).
Publication proved difficult. The extended version was rejected by at least five publishers and journals; the short paper was submitted to several journals, including Science and Nature, with similar results. However, following a visit to Chile by Heinz von Foerster in mid-1973, the paper was further revised and submitted to BioSystems (for which von Foerster was a member of the editorial board), and eventually published in mid-1974 [46]. Much more comprehensive publications have, of course, followed [24,23,44]; but as the first English language publication of the theory of autopoiesis to an international audience, the 1974 BioSystems paper remains a key reference, which continues to be widely read and cited. Further, as the first application of agent-based computer modelling to demarcating the living from the non-living state, it occupies a seminal position in the pre-history of what we now call Artificial Life.
The original computational model of autopoiesis was presented in the form of a detailed, natural language, algorithm [46, Appendix]. This algorithm is reviewed and critiqued, in detail, in [27]. I will return to that critique in due course, but first it is important to have a general, qualitative, view of the model, and the phenomenology it is intended to exhibit.
The chemistry takes places in a discrete, two dimensional, space. Each position in the space is either empty or occupied by a single particle. Particles generally move in random walks in the space. There are three distinct particle types, engaging in three distinct reactions (see Figure 1):
Chains of L particles are permeable to S particles but impermeable to K and L particles. Thus a closed chain, or membrane, which encloses K or L particles effectively traps such particles.
The basic autopoietic phenomenon predicted for this system is the possibility of realising dynamic cell-like structures which, on an ongoing basis, produce the conditions for their own maintenance. Such a system would consist of a closed chain (membrane) of L particles enclosing one or more K particles. Because S particles can permeate through the membrane, there can be ongoing production of L particles. Since these cannot escape from the membrane, this will result in the build up of a relatively high concentration of L particles. On an ongoing basis, the membrane will rupture as a result of disintegration of component L particles. Because of the high concentration of L particles inside the membrane, there should be a high probability that one of these will drift to the rupture site and effect a repair, before the K particle(s) escape, thus re-establishing precisely the conditions allowing the build up and maintenance of that high concentration of L particles.
A secondary phenomenon which may arise is the spontaneous establishment of an autopoietic system from a randomised initial arrangement of the particles.
Both these phenomena were reported and illustrated in the original paper.
One specific weakness of this original model deserves mention here. This is that the K component is not itself produced by any reaction in the putatively autopoietic system. Prima facie this appears to violate the demand for closure in the processes of production. The paper itself is somewhat confusing on this point. A ``six-point key'' is provided for determining whether any given entity is autopoietic. Point 6 of this first appears to require that all components should, indeed, be produced by interactions among the components; but immediately equivocates by allowing some exceptions to this if the relevant components ``participate as necessary permanent constitutive components in the production of other components'' [46, p. 193]. This does appear somewhat clumsy. On the other hand, there must surely be some exceptions allowed (specifically, covering the case of the S particles which are simply harvested from the environment). In any case, as will be discussed later, more recent elaborations of this original model have specifically allowed for production of the K particles [1], so there is no fundamental difficulty here.
Autopoiesis grew and developed in many different directions after 1974. The computational approach was specifically pursued by Milan Zeleny, in a series of publications between 1975 and 1978 [49,54,53,48].
Zeleny first re-implemented the original model in the programming language APL, and then explored a number of different elaborations of this. In particular, he reported such phenomena as growth, change in shape, oscillation in chemical activity, and self-reproduction of autopoietic entities [49,48].
Prima facie these represent important demonstrations of progressively richer, life-like, phenomena within the framework of computational autopoiesis. However, there are some serious difficulties in assessing the significance of these results. The papers cited above do not detail the exact modifications of the original algorithm that gave rise to the new phenomena; but the qualitative descriptions suggest that, in at least some cases, the original constraints of local interaction, random motion of particles, and time-independent particle dynamics, may have been rather arbitrarily relaxed. If that were so, the interest of these results would be seriously diminished--in the sense that the more complex ``macroscopic'' phenomena may have been, in effect, implicitly programmed into individual microscopic particles.
The ideal way to address these questions would be, of course, to directly examine the software responsible. Indeed, this software was made available for use by interested researchers at the time [50, Note 11], although it is not entirely clear whether this was in a form that allowed access to inspect the source code. In any case, and unfortunately, it appears that no copies of this software have survived.1It is therefore now impossible to come to any more definitive conclusions. However, this very impasse clearly suggests an implicit lesson for the methodology of scientific research where the phenomena under investigation have their primary realization in the form of computers executing specific programmes. I will return to this important methodological question below.
Maturana and Varela themselves continued to explore the general paradigm of autopoiesis, of course. Milestones included the publication of Autopoiesis and Cognition: The Realization of the Living in 1980 [23], and the more popular or accessible account, The Tree of Knowledge: The Biological Roots of Human Understanding in 1987 [24]. Also significant was the appearance, in the early 1980's, of two collections of papers by diverse authors, both edited by Milan Zeleny [52,51]. These demonstrated a broadening interest in autopoiesis from researchers in a range of fields.
In the late 1980's there was also a return to the question of a minimal model of (molecular) autopoiesis--but this time not via computation but through real, ``wet'', chemistry. This direction of research was initially introduced in 1988, in a collaboration between Varela and Luisi [20], and quite quickly led to the exhibition of concrete chemical models [19].
Over that same period, from about 1988 to 1992, a strong philosophical interest, and critique, of autopoietic ideas emerged in the literature of general systems theory. This was particularly associated with the work of Fleischaker [4,5,6]. A special issue of the Journal of General Systems, edited by Fleischaker in 1992 [7], provided a forum for a robust debate, from a variety of specialists, over the scope, and, indeed, precise meaning, of autopoiesis.
For my purposes here, the most significant aspect of this discussion is the question of possible ``spaces'', or ``domains'', in which autopoiesis can be realised--and especially the status of purely computational implementations. Fleischaker has the merit of taking a very clear position on this: she holds that, as least insofar as autopoiesis is taken to be definitional or criterial of ``life'', then it must be interpreted as restricted ``entirely to the physical domain'' [5, p. 43]. In turn I take her to mean that computational models of autopoiesis must be regarded only as ``simulations'', and not ``realisations'' of life.
I am not fully persuaded by this position. In particular, it seems to me that we should distinguish between a purely formal description of a computation (e.g., the original ``algorithm'' of Varela et al. [46], or even a specific programme implementing this algorithm) and a concrete, physical, computer which is actually executing such an algorithm. The latter is clearly a very different kind of system from a conventional chemical, or molecular one; but, equally, it is certainly still a physical one. If it hosts autopoietic entities--albeit removed by a significant number of ``levels'' from the underlying ``hardware''--then those entities are surely also still physical. Thus, I would find it hard to categorise such an entity as ``not-living'' solely on a putative basis of its being ``not-physical''.
On the other hand, I am also unconvinced that debate on this issue will be particularly fruitful in any case. I would argue that--within the field of Artificial Life at least--the substantive issue at the current time is not whether a computationally-based autopoietic entity is ``really'' alive, but rather whether we can exhibit computationally-based autopoietic entities with substantially richer, more life-like, phenomenology. The early work of Zeleny was arguably developing in this direction; but, as we have seen, the concrete implementations of that work are not longer available, and it is now very difficult to critically appraise its substance.
I will therefore now return to consider a second phase of (attempted) elaboration of the phenomenology of computational autopoiesis.
As far as I am aware, following Zeleny's initial explorations, little further substantive development of computational autopoiesis was published between c. 1980 and the early 1990's.
As recounted in the preamble, my personal interest began around 1991. I was then completing a detailed re-evaluation of von Neumann's original work on ``self-reproducing'' automata. My conclusion from that study was that this could be best understood as focussed not on self-reproduction per se, but on self-reproduction as a mechanism for the evolutionary growth of complexity [29]. Indeed, von Neumann had demonstrated how the architecture of a relatively passive genetic ``description tape'' coupled with a general purpose, programmable, ``constructor'', would give rise to a dense network of mutational pathways connecting relatively simple automata to arbitrarily complex ones. This is a very significant result; but it does not, in itself, suffice to realise the open-ended evolutionary growth of complexity in artificial systems.
The critical obstacle (one recognised clearly by von Neumann himself) is that his automata are extremely fragile. While they are logically capable of self-reproduction, they can do so only in a completely empty environment, with no disturbance to their operation. In practice, of course, Darwinian evolution could only happen within ecosystems where automata interact both with each other and with a more or less complex environment. Thus, to have a serious prospect of open-ended evolutionary growth of complexity, one requires automata which both have a von Neumann style genetic architecture and are also capable of surviving--maintaining and repairing themselves--in the face of environmental perturbation. Indeed, they should do this even while being open to turnover of their components (the latter being necessary to harvest materials for self-reproduction, if for no other reason).
Now von Neumann had exhibited--in detail--how to realize automata with his genetic architecture within a comparatively simple, two dimensional, discrete space. Similarly, Varela et. al. had exhibited how to realize automata capable of basic self-repair and turnover of components in a similar space (or, at least, one explicitly inspired by von Neumann's). The question therefore arises whether one can engineer a single artificial universe, hopefully of not much greater inherent complexity, in which these two kinds of automaton can be unified.
This then was the problem which I started to discuss with Francisco Varela in the early 1990's [25]. My starting point was the original computational model of autopoiesis. The research programme was, initially at least, rather simple: re-implement that model, demonstrating the original, minimal, autopoietic phenomenology; and then explore elaborations (more particle types, more reactions, etc.) in the general direction of trying to unite the autopoietic functionality with something like a von Neumann genetic system.
However, this programme failed at the very first hurdle: i.e., re-creating the original, minimal, autopoietic phenomenology. It took some time, and several different attempts (which became intermittent, over a number of years), but it gradually became clear that the difficulties were more than superficial.
Eventually, in the Autumn of 1996 I had the opportunity (with support from Chris Langton at the Santa Fe Institute) to make a concentrated study of the problem. This resulted in a detailed analysis of a number of anomalies in the original BioSystems paper [46]. These encompassed both difficulties in interpreting the algorithm, and apparent inconsistencies between the algorithm and the presented experimental results.
While a significant number of detailed issues with the early computational model were uncovered in this process, the great majority of these were relatively minor, and could have been ignored or easily corrected. However, there was one particular phenomenon which was more serious, and which was responsible for the fundamental difficulty in re-creating the original autopoietic phenomena. This is the problem of premature bonding of the L particles. According to the original algorithm, once L particles form they can spontaneously bond together; and, further, once bonded, they become immobile. But this is a fatal flaw for the supposed autopoietic closure of the reaction network: the free L particles are effectively sequestered by the side-reaction of spontaneous bonding. This both makes them essentially unavailable for membrane repair, and also clogs up the interior of the cell, eventually blocking further production.
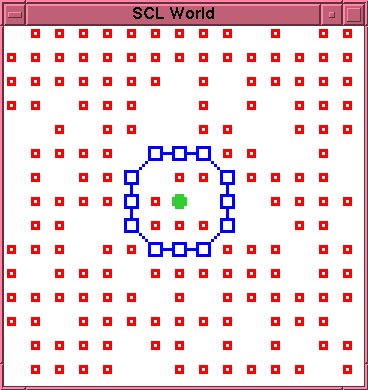
Figures 1 and 2 show an experimental example of this. Figure 1 shows the initial configuration of an externally created cell. Figure 2 shows the state of the cell after 110 time steps. The membrane has not yet suffered any decay. However, the interior of the membrane is now completely clogged with bonded--and thus immobile--L particles. Only two open positions remain inside the membrane, one occupied by the K particle. Since the production reaction requires two S particles adjacent to each other and to the K particle, there is no longer any available site for further production within the membrane, and further production of L particles is impossible. It follows that, whenever the membrane does eventually rupture, there will be no mobile L particles available to effect a repair.
This is a very robust failure. It is not sensitive to any precise details of the implementation. Indeed, the same problem was retrospectively identified in two other, independent, attempts at re-implementation [15,34]; and may well have been present in the Zeleny work also (but superficially masked there through the introduction of non-local interactions etc.).
This then provided the basis for a more extended and detailed discussion with Francisco Varela. But, of course, there had been a long lapse of time since the original experimental work had been carried out, c. 1971. Further, it should be remembered that the work had taken place against a backdrop of great turbulence in Chile. Varela had been a prominent supporter of the Allende government. Following the coup d'etat and the coming to power of Pinochet in September 1973, Varela, like many others, was effectively forced into exile [45]. These events co-incided directly with the development of the BioSystems paper. Circumstances had therefore combined to make it very difficult to recall or reconstruct technical details of that work, some 25 years later.
This was the situation of the discussion in late 1996, when chance intervened. Varela was now based in Paris, and he there happened to receive a shipment of papers and files, apparently belonging to him, which had just been located back at the University of Chile. Among these papers, he identified several contemporary, albeit fragmentary, records of the early computational exploration of autopoiesis. Critically, these included an early discursive description of the model and a listing of a version of the actual computer code (in FORTRAN IV).
With the help of these documents it was finally possible to identify, with reasonable confidence, exactly the causes of the discrepancies which had been identified in the BioSystems paper.2 In particular, an explanation was found for the control of premature bonding in the original model. It transpired that the model had actually included an additional, undocumented, interaction: chain-based bond inhibition. The effect of this was that free L particles in the immediate vicinity of the membrane (or, indeed, any chain of L particles) were inhibited from spontaneously forming bonds with each other. Provided the cell is not very big, this is sufficient to prevent premature bonding and ensure a continuing supply of free, mobile, L particles to repair membrane ruptures as and when they arise. Once this interaction was re-introduced, the original autopoietic phenomenology of ongoing self-repair, as reported in [46], was easily realised again. Detailed experiments documenting and demonstrating this were presented at ECAL 1997 [33].
The implicit limitation of the chain-based bond inhibition mechanism to comparatively small cells should be carefully noted: it reveals, in particular, that this original form of autopoietic cell will not easily support growth, or, more particularly, cell reproduction by fission. This is obviously rather important for any projected extension to realising Darwinian evolution of such cells. We will return to this limitation below.
While this eventual re-creation of the original autopoietic phenomenology ultimately hinged on a rather mundane technical refinement, I would argue that it was still a useful, if modest, contribution, for two distinct reasons:
It should be admitted that, in contrast to the other interactions in this model, chain-based bond inhibition has a distinctly ad hoc flavour. That being the case, it can reasonably be asked why this interaction should be considered as being ``acceptable''--that is, as not detracting from the significance of the reported phenomena. This is particularly moot, because certain other (conjectured) interactions in the later models of Zeleny were treated more critically above (section 4). However, it seems to me that a distinction should be drawn here between interactions that are ad hoc simply in the sense of ``unfamiliar'' or ``unphysical'', and interactions that are non-local (in some relevant ``space'') or that have pre-programmed time-variation. As already noted, the latter have the clear potential to allow both aggregate and time-structured phenomena to be effectively ``programmed in'' (either in single agents, or in the environment as a whole). For example, apparently macroscopic ``cellular'' organisation might, in effect, be an artefact constructed and co-ordinated by a single ``central control'' agent; in which case, its ``emergence'' would presumably be of little interest.
By contrast, chain-based bond inhibition is still a perfectly local, and time-independent, interaction, occurring on a smaller scale than, and un-influenced by, macroscopic cellular organisation. It is for this reason that, in my view, the identification of this additional interaction does not undermine the significance of the dynamic, macroscopic, cellular organisation originally reported for this model.
Of course, the question at stake here--of choosing appropriate abstractions or ``axiomatizations'' of primitive agents--is a difficult one, which has been at the heart of the field of Artificial Life right since its inception. Indeed, in a lecture originally dating from 1949, von Neumann captured this tension explicitly and succinctly:
... one may define parts in such numbers, and each of them so large and involved, that one has defined the whole problem away. If you choose to define as elementary objects things which are analogous to whole living organisms, then you obviously have killed the problem, because you would have to attribute to these parts just those functions of the living organism which you would like to describe or to understand. So, by choosing the parts too large, by attributing too many and too complex functions to them, you lose the problem at the moment of defining it. [47, p. 76]
This continues to be a matter of debate in the field, and I do not suggest that the particular demarcation I have suggested above--based specifically on ``locality'' and ``time-independence''--offers anything more than a very rough, heuristic, guide. Particular models, and their associated phenomenology, must still be critically evaluated on their individual merits.
In closing this discussion of the clarification of the original model of computational autopoiesis it is, perhaps, equally important to comment on something that this work did not demonstrate: namely, it certainly did not show that there was, or is, any obstacle of principle in realising autopoietic organisation in an artificial, computational, medium. This must be stated clearly, as precisely an opposite interpretation has recently been attributed to this work:
... The non-computability of Autopoietic systems, as advanced here, apparently collides with the simulation results involving tessellation automatas (sic) [46]. But new versions of this simulation show that the original report of computational autopoiesis was flawed, as it used a non-documented feature involving chain-based bond inhibition [33]. Thus the closure exhibited by tessellation automatas is not a consequence of the ``network'' of simulated processes, but rather an artifact of coding procedures [33].
--Letelier et al. [18, p. 270]
For context, the over-arching conjecture being advanced by Letelier et al. is that autopoiesis is, in some fundamental sense, a ``non-computable'' process. By this they mean that while it can (obviously?) be realised in real physico-chemical form, there is some essential aspect of this process that cannot, even in principle, be realised in a strictly computational system. It amounts to a specific denial of the Church-Turing thesis (regarding the ultimate computability of the entire physical universe). But clearly there is a logical conflict between such a conjecture and the original illustration of the very concept of autopoiesis via a perfectly computational model. The quotation above is an attempt to resolve this conflict.
However, this is a most peculiar analysis.
It is true, of course, that the original report of computational autopoiesis was ``flawed'', and, indeed, that this was primarily because ``it used a non-documented feature involving chain-based bond inhibition''. However, the citation for the last sentence in this quotation is simply wrong: there is no reference whatsoever in [33] to any distinction between ``simulated processes'' and ``coding procedures''. Nor can I conceive, even now, in retrospect, of any basis for assigning a different status to ``chain-based bond inhibition'' compared to any other interaction in that particular artificial chemistry (production, bonding, absorption, etc.).
Thus, there is no basis whatever for supposing that this clarification of the ``original report'' undermines the essential result. The only flaw was in enumerating the processes or interactions underlying them: one necessary interaction was omitted from this description. The phenomena originally reported were accurate, and ultimately reproducible in perfectly ``computable'' form. If it is accepted that the phenomena reported in the original paper constituted ``autopoiesis''--as it apparently is by Letelier et al.--then they still constitute autopoiesis. This was stated most explicitly in the conclusion of [33]:
It should be emphasised that the substantive point of this paper is to correct the historical record. ... However, this correction does not add to, or modify, the original conceptual foundation of autopoiesis in any significant way.
I conclude that the overall thesis of Letelier et al.--of the ``non-computability'' of autopoietic systems--should be taken as refuted, rather than corroborated, by the results of [33].
I have critically reviewed two distinct bodies of work in computational autopoiesis above--the original model of Varela et. al., and the developments of that by Zeleny. These are certain similarities in my comments. In both cases I have identified anomalies or difficulties of interpretation of the published results. However, the final conclusions are quite different. In the case of the original model, it proved possible to locate the source code (or, at least, a close relative) on which the results were based; as a result, the substantive difficulties were fully resolved. For Zeleny's work, unfortunately, the source code is apparently lost; definitive resolution of questions relating to it, at this stage, is therefore extremely difficult, if not impossible.
I should emphasize that these differing outcomes are largely due to chance--the fortuitous re-discovery of the Varela documents--rather than design. However, regardless of that, they suggest a methodological lesson worth digesting. At the time of the original publications considered above, the technological facilities were not generally available to support easy distribution or access to accompanying code--but this is no longer the case. I would suggest therefore that as a general principle, published reports on computer models of ALife should be accompanied by access to the program code for the models on the World Wide Web; and indeed, that practice has now been followed in the cases of the surviving documents from Varela et. al. [27] and the successful modern re-implementation [28,33].
As already explained, a natural research programme in computational autopoiesis is to attempt to realise Darwinian evolution among lineages of autopoietic cells. This requires--among other things--the realisation of cells that are at least capable of self-reproduction through simple growth and fission.
In pursuing this programme, an initial problem which arises then is individuation.
My own exploration of this problem [30] begins with a conceptual comparison of autopoiesis and Stuart Kauffman's idea of a ``collectively autocatalytic set'' [16]. I conclude that there is a critical distinction between these precisely in that autopoiesis requires self-generated ``individuation''. This is classically captured by stating that the autopoietic network of processes must give rise to a boundary; however, it turns out that it is not entirely clear what should qualify, in general, as a ``boundary''. In [30] I suggest a heuristic test for this, which essentially asks whether two putatively individual cells, in direct contact with each other, can reliably maintain their separate identities.
This test is, of course, motivated directly by the issue of self-reproduction. In the situation of self-reproduction of an autopoietic cell by growth and fission, then the parent and offspring will clearly initially be in direct proximity to each other. If they fail to maintain their separate identities--if they can as easily merge back into one cell--this would hardly qualify as ``self-reproduction'' in the functional sense required for evolution, as such merging would undermine the very Malthusian population growth required for Darwinian selection.3 This then is precisely the acid test for individuation: whether an entity can maintain its self-identity when it interacts with another entity of exactly the same material components and organisation.
The test was first applied to a variety of ``classical''
artificial life systems: the
![]() -universes
[14],
Alchemy
[8,9], and
Tierra [42]. In these cases, the
conclusion was as expected: it served to make more precise the
a priori judgement that none of these should be considered
as realising computational autopoiesis.
-universes
[14],
Alchemy
[8,9], and
Tierra [42]. In these cases, the
conclusion was as expected: it served to make more precise the
a priori judgement that none of these should be considered
as realising computational autopoiesis.
The surprise came when the test was applied to the original (re-implemented) model of computational autopoiesis [46,33]. Though experimental results were not presented in detail, it appears that the chain-based bond inhibition reaction, while essential to the self-repair of a single isolated cell, also has the unintended side effect of inhibiting maintenance of the bounding membrane when two membranes are adjacent to each other. This means that, if anything, adjacent agents tend positively to merge rather than to maintain their individuality. In turn, if the proposed heuristic test is accepted as an operational test for autopoietic individuation, we are forced to the somewhat controversial conclusion that the original minimal model, which was intended precisely to be an exemplar of the concept, is not in fact properly autopoietic.
However, to pre-empt mis-interpretation, I would emphasise that this result still does not argue for any obstacle in principle to computational autopoiesis. It merely demonstrates that robust autopoietic organisation is not easy or trivial to achieve.
Two direct elaborations of the original model of computational autopoiesis have been published more recently.
Breyer et al. [1] first relaxed the original restrictions on the motion of bound L particles to allow the formation of flexible chains and, indeed, membranes. Allowing multiple, doubly-bonded, L particles to occupy a single lattice site further enhanced membrane motion. Adopting more sophisticated ``bond re-arrangement'' interactions then allows chain fragments formed within the cell to be dynamically integrated into the membrane. These mechanisms together obviate the need for the chain-based bond inhibition mechanisms--since now, even if L particles spontaneously bond in the interior of the cells, the oligomers so formed remain mobile, and through bond rearrangement, can still successfully function to repair membrane ruptures. In this way, the model is reported as successfully supporting cell growth.
The authors separately consider mechanisms for production of further K particles within a cell. As previously noted, this possibility was absent in the original model, but is, of course, a necessary condition for any development toward self-reproduction. However, the full achievement of self-reproduction is not reported.
Rather similar developments were independently reported by McMullin and Groß [31]. Again, bound L particles were allowed to move, facilitating flexible membranes; a somewhat more complex bond re-arrangement interaction was introduced to facilitate cell growth; and the chain-based bond inhibition interaction was discarded. In this case, however, instead of simply permitting spontaneous L bonding within the cell (and then relying on mobility of these fragments to sustain repair), chain initiation (i.e., bonding between two free L particles) was disabled. This meant that the interior of the cell could still sustain a relatively large concentration of highly mobile free L particles. Other refinements included ``smart'' repair of the membrane (where membrane particles were replaced as soon as they started to decay--even before the membrane was actually ruptured) and ``membrane affinity'' of free L particles. The latter meant that the membrane tended to take a bi-layer form; the outer layer particles being bonded, and the inner layer providing a stock of free L particles to effect very efficient repair.
These modifications together permitted the establishment of autopoietic cells with much greater dynamic stability (longer lifetimes) and/or cells capable of sustained growth. However, it should be noted that, with chain initiation disabled, this particular model would not be capable of exhibiting spontaneous cell generation de novo. Again, this paper identifies the enriched phenomenology as potential steps toward the realisation of self-reproduction of autopoietic cells, through growth and fission; but does not report the achievement of that.
A significantly different approach to computational autopoiesis, termed Lattice Artificial Chemistry (LAC) has been recently pioneered in a series of studies by Ono and Ikegami.
LAC differs from the original model of Varela et al. [46] (and its direct descendants above) in adopting a coarse grained approach. In the original model, each lattice site can generally be occupied by at most one particle: it is fine-grained to the single molecule level. In LAC, by contrast, each single lattice site is permitted to host many particles, of many distinct molecular species.
Using this approach Ono and Ikegami have progressively reported:
Because LAC uses the coarse grained approach, direct comparisons with the fine-grained models are difficult. For example, when multiple cells are present in the same space, they normally share membranes, so that a population of cells exhibits a ``honeycomb'' structure. It is difficult to assess the degree of ``individuation'' on the part of single cells in this case (as multiple cells contribute to the maintenance of any given boundary).
The exhibition of selection at the cell level seems particularly significant [39]. At this point, this is still limited to selection among a pre-designed variety of cell strains--there is nothing like a von Neumann style genetic architecture, or programmable constructor. Nonetheless, the LAC methodology clearly presents a very fruitful avenue of research.
While this paper has concentrated on the development of computational autopoiesis, per se, this is clearly related to several other, parallel, developments.
The Chemoton of Tibor Gánti [10], though apparently developed quite independently, is clearly related to cellular autopoiesis; indeed, this first (English) report on the chemoton concept appeared in the same journal, BioSystems, as the first presentation of Autopoiesis [46], but in the following year (1975). These parallels--and links to a wide variety of much earlier work--have been more recently reviewed by Gánti [11].
The chemoton is again a proposal for an abstract, ``minimal'' cell, consisting of a collectively autocatalytic network of reactions which is enclosed within a membrane, which is also generated and maintained by the reaction network. It differs from the minimal autopoiesis model in explicitly including a ``genetic subsystem''. It is also rather more detailed in its analysis of the required chemical dynamics (kinetics etc.), and aims at supporting self-reproduction by growth and fission even in the minimal version.
While the provision of a genetic system is significant, it should be emphasised that, in the basic chemoton, this is limited to replication of varieties of ``polymers'' which then have direct chemical effects. The polymers do not participate in a translation process; thus, this subsystem is again relatively limited in scope compared to a von Neumann style genetic architecture, or programmable constructor.
The chemoton has been subject to various computer simulation studies, particularly by Csendes [3]. However, these were based essentially on an Ordinary Differential Equation approach, rather than being molecular or agent-based. This means that, in particular, the distributed, spatial, dynamics of membrane growth, fission, and individuation, were not substantively modelled.
Other computational systems with some connection or similarity to
autopoiesis include Alchemy
[8,9], the
![]() -universes
[14], Coreworld
[41] and Tierra
[42]. However, as already mentioned,
while these do involve closed, self-sustaining, reaction
networks, they do not have mechanisms for the self-generation of
spatial boundaries.
-universes
[14], Coreworld
[41] and Tierra
[42]. However, as already mentioned,
while these do involve closed, self-sustaining, reaction
networks, they do not have mechanisms for the self-generation of
spatial boundaries.
Another very interesting line of work involves much more physico-chemically realistic computer models of minimal cellular structures. Rasmussen and colleagues, in particular, have recently exhibited spontaneous emergence of such hierarchical levels of organisation [40]. This line of attack poses some scaling difficulties, as the computational demands of physico-chemical realism rise very rapidly. On the other hand, the cost of computation continues to fall, so this may well be a very fruitful domain of further research in the immediate future.
Finally, we should note that there are clear conceptual connections between the idea of autopoiesis and Robert Rosen's metabolism-repair systems and ``closure under efficient causation'' [43]. This connection has recently been made explicit by Letelier et al. [18], with the concrete suggestion that Rosen's work may provide an appropriate formal framework for the understanding of autopoiesis. A similar conjecture has also been recently presented by Nomura [35].
This approach immediately encounters the difficulty that Rosen explicitly argued that his form of closure could not, even in principle, be embedded within a purely computational system. By contrast, computer realisation has been an explicit exemplar of autopoietic closure from the very start. As already discussed, Letelier et al. [18] have attempted to resolve this by interpreting [33] as showing that computational autopoiesis was fundamentally flawed from the beginning; but this is not an interpretation I can share. Accordingly, at this point, while I would agree that Rosen's work presents an interesting avenue for further exploration and contrast with autopoiesis, it is certainly not yet demonstrated than it can contribute a substantive, formal, theory to underpin it.
The idea of autopoiesis, and especially its computational realisation, has proved very fruitful in its development and elaboration over the last thirty years. As we have seen, it represents a core thread in the history of what we now call Artificial Life, linking back directly to von Neumann's founding work in evolutionary automata.
We now have a comparatively clear formulation of several key open problems. One very general challenge is to articulate the relationship between the ``fine-grained'' and ``coarse-grained'' models of computational autopoiesis, and the relationship between both of these and work in ``wet'' artificial life. More specific, concrete challenges revolve around the exhibition of substantive Darwinian evolutionary phenomena among lineages of artificial (computational) autopoietic individuals. Phenomena of particular interest would include the evolution of individuality itself [2], the exhibition of a molecular-agent computational model of the Chemoton [11], and the evolution of von Neumann-style genetic architecture based on a programmable constructor [29].
Francisco Varela was a pivotal figure in the development of our field to this point. His influence and intellectual legacy provides a strong foundation to tackle these difficult and profound problems that we still face in properly understanding the organisation of the living, its origin and its evolution.
Financial support for the work has been provided by the Research Institute for Networks and Communications Engineering (RINCE) at Dublin City University.
 |
| DCU Alife Laboratory | |
| Research Institute for Networks and Communications Engineering (RINCE) | |
| Dublin City University | |
| Dublin 9 | |
| Ireland. | |
| Voice: | +353-1-700-5432 |
|---|---|
| Fax: | +353-1-700-5508 |
| E-mail: | Barry.McMullin@dcu.ie |
| Web: | http://www.eeng.dcu.ie/~mcmullin |
The resources comprising this paper are retrievable in various formats via:
©
2004 The MIT Press
The final version of this article has been accepted for publication in Artificial Life, Vol. 10, Issue 3, Summer 2004. Artificial Life is published by The MIT Press.
Certain rights have been reserved to the author, according to the relevant MIT Press Author Publication Agreement.
Copyright © 2004 All Rights Reserved.
Timestamp: 2004-06-14